Abstract
Research Article
Sites and Zones of Maximum Reactivity of the most Stable Structure of the Receptor-binding Domain of Wild-type SARS-CoV-2 Spike Protein: A Quantum Density Functional Theory Study
Ernesto López-Chávez*, Alberto García-Quiroz, Yesica Antonia Peña-Castañeda, José Antonio Irán Díaz-Góngora, José Alberto Mendoza-Espinoza, Jose Antonio López-Barrera and Fray de Landa Castillo-Alvarado
Published: 12 April, 2024 | Volume 9 - Issue 1 | Pages: 008-016
Today, it is well known that Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has four types of proteins within its structure, between them the spike protein (S). The infection mechanism is carried out by the entry of the virus into the human host cell through the S protein, which strongly interacts with the human cell receptor angiotensin-converting enzyme 2 (ACE2). In this work, we propose an atomic model of the Receptor Binding Domain (RBD) of the S spike protein of the wild-type SARS-CoV-2 virus. The molecular structure of the model was composed of 50 amino acids that were chemically bonded, starting with Leucine and ending with one amino acid Tyrosine. The novelty of our work lies in the importance of knowing the sites and zones of maximum reactivity of the RBD from the fundamental levels of quantum mechanics considering the atomic structure of matter. For this, the local and global reactivity indices of the RBD were calculated, such as frontier orbitals, Highest Occupied Molecular Orbital (HOMO) and Lowest Unoccupied Molecular Orbital (LUMO), Fukui indices, chemical potential, chemical hardness, electrophilicity index; with this, it will be possible to know what type of molecules are more likely to interact with the RBD structure, and in this way, new knowledge will be generated at the quantum, atomic and molecular level to inhibit the virulent effects of wild-type SARS-CoV-2. Finally, in order to identify the functional groups within the most stable structure and thereby verify the future reactions that can be carried out between the RBD structure and biomolecules, the Infrared (IR) absorption spectrum was calculated. For this work, we used Material Studio v6.0 which uses the density functional theory (DFT) implemented in its DMol3 computational code. The IR spectrum was obtained using the Spartan ‘94 computer code. One novelty would be that we found nine amino acids more that could make the RBD and ACE2 binding further the already known. Thus, the Mulliken charge distribution indicates that the highest concentrations of positive and negative charge are found in the zones 477S, 478T, 484E, and 501N amino acids letting ionic or Van der Waals possible interactions with other structures.
Read Full Article HTML DOI: 10.29328/journal.jcicm.1001047 Cite this Article Read Full Article PDF
Keywords:
Highest occupied molecular orbital; Lowest unoccupied molecular orbital; Density functional theory; Receptor binding domain; Severe acute respiratory syndrome coronavirus 2; Corona virus disease of 2019
References
- Yan ZP, Yang M, Lai CL. COVID-19 Vaccines: A Review of the Safety and Efficacy of Current Clinical Trials. Pharmaceuticals (Basel). 2021 Apr 25;14(5):406. doi: 10.3390/ph14050406. PMID: 33923054; PMCID: PMC8144958.
- Hu B, Guo H, Zhou P, Shi ZL. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021 Mar;19(3):141-154. doi: 10.1038/s41579-020-00459-7. Epub 2020 Oct 6. Erratum in: Nat Rev Microbiol. 2022 May;20(5):315. PMID: 33024307; PMCID: PMC7537588.
- V'kovski P, Kratzel A, Steiner S, Stalder H, Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021 Mar;19(3):155-170. doi: 10.1038/s41579-020-00468-6. Epub 2020 Oct 28. PMID: 33116300; PMCID: PMC7592455.
- Khare P, Sahu U, Pandey SC, Samant M. Current approaches for target-specific drug discovery using natural compounds against SARS-CoV-2 infection. Virus Res. 2020 Dec;290:198169. doi: 10.1016/j.virusres.2020.198169. Epub 2020 Sep 24. PMID: 32979476; PMCID: PMC7513916.
- Jawad B, Adhikari P, Podgornik R, Ching WY. Key Interacting Residues between RBD of SARS-CoV-2 and ACE2 Receptor: Combination of Molecular Dynamics Simulation and Density Functional Calculation. J Chem Inf Model. 2021 Sep 27;61(9):4425-4441. doi: 10.1021/acs.jcim.1c00560. Epub 2021 Aug 24. PMID: 34428371.
- Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020 Apr 16;181(2):281-292.e6. doi: 10.1016/j.cell.2020.02.058. Epub 2020 Mar 9. Erratum in: Cell. 2020 Dec 10;183(6):1735. PMID: 32155444; PMCID: PMC7102599.
- Kang YF, Sun C, Zhuang Z, Yuan RY, Zheng Q, Li JP, Zhou PP, Chen XC, Liu Z, Zhang X, Yu XH, Kong XW, Zhu QY, Zhong Q, Xu M, Zhong NS, Zeng YX, Feng GK, Ke C, Zhao JC, Zeng MS. Rapid Development of SARS-CoV-2 Spike Protein Receptor-Binding Domain Self-Assembled Nanoparticle Vaccine Candidates. ACS Nano. 2021 Feb 23;15(2):2738-2752. doi: 10.1021/acsnano.0c08379. Epub 2021 Jan 19. PMID: 33464829.
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 May;581(7807):215-220. doi: 10.1038/s41586-020-2180-5. Epub 2020 Mar 30. PMID: 32225176.
- Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI; Sheffield COVID-19 Genomics Group; McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020 Aug 20;182(4):812-827.e19. doi: 10.1016/j.cell.2020.06.043. Epub 2020 Jul 3. PMID: 32697968; PMCID: PMC7332439.
- Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 2022 Jan;23(1):3-20. doi: 10.1038/s41580-021-00418-x. Epub 2021 Oct 5. PMID: 34611326; PMCID: PMC8491763.
- Yamasoba D, Kimura I, Nasser H, Morioka Y, Nao N, Ito J, Uriu K, Tsuda M, Zahradnik J, Shirakawa K, Suzuki R, Kishimoto M, Kosugi Y, Kobiyama K, Hara T, Toyoda M, Tanaka YL, Butlertanaka EP, Shimizu R, Ito H, Wang L, Oda Y, Orba Y, Sasaki M, Nagata K, Yoshimatsu K, Asakura H, Nagashima M, Sadamasu K, Yoshimura K, Kuramochi J, Seki M, Fujiki R, Kaneda A, Shimada T, Nakada TA, Sakao S, Suzuki T, Ueno T, Takaori-Kondo A, Ishii KJ, Schreiber G; Genotype to Phenotype Japan (G2P-Japan) Consortium; Sawa H, Saito A, Irie T, Tanaka S, Matsuno K, Fukuhara T, Ikeda T, Sato K. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell. 2022 Jun 9;185(12):2103-2115.e19. doi: 10.1016/j.cell.2022.04.035. Epub 2022 May 2. PMID: 35568035; PMCID: PMC9057982.
- Qiao B, Olvera de la Cruz M. Enhanced Binding of SARS-CoV-2 Spike Protein to Receptor by Distal Polybasic Cleavage Sites. ACS Nano. 2020 Aug 25;14(8):10616-10623. doi: 10.1021/acsnano.0c04798. Epub 2020 Aug 4. PMID: 32806067.
- Gao K, Wang R, Chen J, Cheng L, Frishcosy J, Huzumi Y, Qiu Y, Schluckbier T, Wei X, Wei GW. Methodology-Centered Review of Molecular Modeling, Simulation, and Prediction of SARS-CoV-2. Chem Rev. 2022 Jul 13;122(13):11287-11368. doi: 10.1021/acs.chemrev.1c00965. Epub 2022 May 20. PMID: 35594413; PMCID: PMC9159519.
- Wang Q, Wang L, Zhang Y, Zhang X, Zhang L, Shang W, Bai F. Probing the Allosteric Inhibition Mechanism of a Spike Protein Using Molecular Dynamics Simulations and Active Compound Identifications. J Med Chem. 2022 Feb 24;65(4):2827-2835. doi: 10.1021/acs.jmedchem.1c00320. Epub 2021 Aug 20. PMID: 34415156.
- Padhi AK, Rath SL, Tripathi T. Accelerating COVID-19 Research Using Molecular Dynamics Simulation. J Phys Chem B. 2021 Aug 19;125(32):9078-9091. doi: 10.1021/acs.jpcb.1c04556. Epub 2021 Jul 28. PMID: 34319118.
- Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. The proximal origin of SARS-CoV-2. Nat Med. 2020 Apr;26(4):450-452. doi: 10.1038/s41591-020-0820-9. PMID: 32284615; PMCID: PMC7095063.
- Vosko SH, Wilk L, Nusair M. Accurate spin-dependent electron liquid correlation energies for local spin density calculations: A critical analysis. Can J Phys. 1980; 58:1200-1211. http://dx.doi.org/10.1139/p80-159
- Perdew JP, Wang Y. Accurate and simple analytic representation of the electron-gas correlation energy. Phys Rev B Condens Matter. 1992 Jun 15;45(23):13244-13249. doi: 10.1103/physrevb.45.13244. PMID: 10001404.
- Becke AD. A multicenter numerical integration scheme for polyatomic molecules. J Chem Phys. 1988; 88:2547-2553 https://doi.org/10.1063/1.454033
- Delley B. Theoretical and Computational Chemistry, 2, 221-254, Volume book: Modern density functional theory: a tool for chemistry, edited by J.M. Seminario, P. Politzer, Elsevier Science: Amsterdam, ISBN 0-444-82171-6. https://doi.org/10.1016/S1380-7323(05)80037-8
- Perdew JP, Burke K, Ernzerhof M. Generalized Gradient Approximation Made Simple. Phys Rev Lett. 1996; 77: 3865. Erratum Phys. Rev. Lett. 78: 1396. https://doi.org/10.1103/PhysRevLett.78.1396
- Qi L, Fa-tang L. Recent advances in molecular oxygen activation via photocatalysis and its application in oxidation reactions. Chem Engi J. 2021; 421: 129915. https://doi.org/10.1016/j.cej.2021.129915
- Noorizadeh S, Taghipour KA. A regioselectivity descriptor based on atomic Weizsäcker kinetic energy. Chem Phys Lett. 2021; 770:138455. https://doi.org/10.1016/j.cplett.2021.138455
- Parr RG, Yang W. Density functional approach to the frontier-electron theory of chemical reactivity. J Am Chem Soc. 1984; 106(14): 4049–4050 https://doi.org/10.1021/ja00326a036
- Parr RG, Yang W. Density Functional Theory of Atoms and Molecules. Oxford University Press, N.Y. USA, 1989; https://doi.org/10.1002/qua.560470107
- Parr RG, Donnelly RA, Levy M, Palke WE. Electronegativity: the density functional viewpoint. J Chem Phys. 1978; 68:3801–3807 http://dx.doi.org/10.1063/1.436185
- Mulliken RS. Electronic population analysis on LCAO-MO molecular wave functions. I and Electronic population analysis on LCAO-MO molecular wave functions. II. Overlap populations, bond orders, and covalent bond energies. J Chem Phys. 1955; 10(23): 1833-1846. https://doi.org/10.1063/1.1740588
- Boretti A. Favipiravir use for SARS CoV-2 infection. Pharmacol Rep. 2020; 72(6): 1542-1552. https://doi.org/10.1007/s43440-020-00175-2.
- Valle C, Martin B, Touret F, Shannon A, Canard B, Guillemot JC, Coutard B, Decroly E. Drugs against SARS-CoV-2: What do we know about their mode of action? Rev Med Virol. 2020 Nov;30(6):1-10. doi: 10.1002/rmv.2143. Epub 2020 Aug 11. PMID: 32779326; PMCID: PMC7435512.
- Xia S, Chen E, Zhang Y. Integrated Molecular Modeling and Machine Learning for Drug Design. J Chem Theory Comput. 2023 Nov 14;19(21):7478-7495. doi: 10.1021/acs.jctc.3c00814. Epub 2023 Oct 26. PMID: 37883810; PMCID: PMC10653122.
- Gupta R, Srivastava D, Sahu M, Tiwari S, Ambasta RK, Kumar P. Artificial intelligence to deep learning: machine intelligence approach for drug discovery. Mol Divers. 2021 Aug;25(3):1315-1360. doi: 10.1007/s11030-021-10217-3. Epub 2021 Apr 12. PMID: 33844136; PMCID: PMC8040371.
Figures:
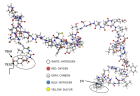
Figure 1

Figure 2
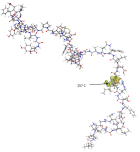
Figure 3

Figure 4
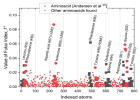
Figure 5
Figure 6
Similar Articles
-
Immunocompromised patients with SARS-CoV-2 infection in intensive care units, outcome and mortalityCynthia DENIS*,Hamid MERDJI,Mathieu BALDACINI,Maleka SCHENCK,Thierry ARTZNER,Yoann GRIMAUD,Thierry LAVIGNE,Ferhat MEZIANI,Vincent CASTELAIN,Raphaël CLERE-JEHL,Francis SCHNEIDER,Guillaume MOREL. Immunocompromised patients with SARS-CoV-2 infection in intensive care units, outcome and mortality. . 2021 doi: 10.29328/journal.jcicm.1001036; 6: 021-025
-
Sites and Zones of Maximum Reactivity of the most Stable Structure of the Receptor-binding Domain of Wild-type SARS-CoV-2 Spike Protein: A Quantum Density Functional Theory StudyErnesto López-Chávez*, Alberto García-Quiroz, Yesica Antonia Peña-Castañeda, José Antonio Irán Díaz-Góngora, José Alberto Mendoza-Espinoza, Jose Antonio López-Barrera, Fray de Landa Castillo-Alvarado. Sites and Zones of Maximum Reactivity of the most Stable Structure of the Receptor-binding Domain of Wild-type SARS-CoV-2 Spike Protein: A Quantum Density Functional Theory Study. . 2024 doi: 10.29328/journal.jcicm.1001047; 9: 008-016
Recently Viewed
-
Prevalence and Risk Factors to Preterm Labor through a Study in Jiblah University Hospital, Ibb, Governorate, YemenAfaf Alsharif*,Zainab Said,Fatima Mokabes,Leena Ameen,Alya Alqadri,Thekra Musaed,Bushra Musaed,Ala’a Ahmed,Halaa Rigih. Prevalence and Risk Factors to Preterm Labor through a Study in Jiblah University Hospital, Ibb, Governorate, Yemen. J Community Med Health Solut. 2025: doi: 10.29328/journal.jcmhs.1001053; 6: 020-026
-
Navigating Neurodegenerative Disorders: A Comprehensive Review of Current and Emerging Therapies for Neurodegenerative DisordersShashikant Kharat*, Sanjana Mali*, Gayatri Korade, Rakhi Gaykar. Navigating Neurodegenerative Disorders: A Comprehensive Review of Current and Emerging Therapies for Neurodegenerative Disorders. J Neurosci Neurol Disord. 2024: doi: 10.29328/journal.jnnd.1001095; 8: 033-046
-
The Color of Diseases and Herbs-chromatic Illustrating the Yin-yang Regulation Theory in Traditional Chinese MedicineHuigang Liu*. The Color of Diseases and Herbs-chromatic Illustrating the Yin-yang Regulation Theory in Traditional Chinese Medicine. Ann Biomed Sci Eng. 2025: doi: 10.29328/journal.abse.1001034; 9: 001-004
-
Analyzing Maternal Inheritance of Mitochondrial DNA using PCR-RFLPNidhi Sharma*,Ruchika Kaushik,Tabin Millo,Chittaranjan Behera. Analyzing Maternal Inheritance of Mitochondrial DNA using PCR-RFLP. J Forensic Sci Res. 2025: doi: 10.29328/journal.jfsr.1001081; 9: 054-058
-
Menstrual Taboos and Child Rights: Death of a Girl during MenarcheVishadha Perera*. Menstrual Taboos and Child Rights: Death of a Girl during Menarche. J Forensic Sci Res. 2025: doi: 10.29328/journal.jfsr.1001080; 9: 050-053
Most Viewed
-
Sinonasal Myxoma Extending into the Orbit in a 4-Year Old: A Case PresentationJulian A Purrinos*, Ramzi Younis. Sinonasal Myxoma Extending into the Orbit in a 4-Year Old: A Case Presentation. Arch Case Rep. 2024 doi: 10.29328/journal.acr.1001099; 8: 075-077
-
Evaluation of Biostimulants Based on Recovered Protein Hydrolysates from Animal By-products as Plant Growth EnhancersH Pérez-Aguilar*, M Lacruz-Asaro, F Arán-Ais. Evaluation of Biostimulants Based on Recovered Protein Hydrolysates from Animal By-products as Plant Growth Enhancers. J Plant Sci Phytopathol. 2023 doi: 10.29328/journal.jpsp.1001104; 7: 042-047
-
Feasibility study of magnetic sensing for detecting single-neuron action potentialsDenis Tonini,Kai Wu,Renata Saha,Jian-Ping Wang*. Feasibility study of magnetic sensing for detecting single-neuron action potentials. Ann Biomed Sci Eng. 2022 doi: 10.29328/journal.abse.1001018; 6: 019-029
-
Physical activity can change the physiological and psychological circumstances during COVID-19 pandemic: A narrative reviewKhashayar Maroufi*. Physical activity can change the physiological and psychological circumstances during COVID-19 pandemic: A narrative review. J Sports Med Ther. 2021 doi: 10.29328/journal.jsmt.1001051; 6: 001-007
-
Pediatric Dysgerminoma: Unveiling a Rare Ovarian TumorFaten Limaiem*, Khalil Saffar, Ahmed Halouani. Pediatric Dysgerminoma: Unveiling a Rare Ovarian Tumor. Arch Case Rep. 2024 doi: 10.29328/journal.acr.1001087; 8: 010-013

HSPI: We're glad you're here. Please click "create a new Query" if you are a new visitor to our website and need further information from us.
If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."